Hermite®
AI4S-Enhanced CADD Tool for Accelerating Rational Drug Discovery
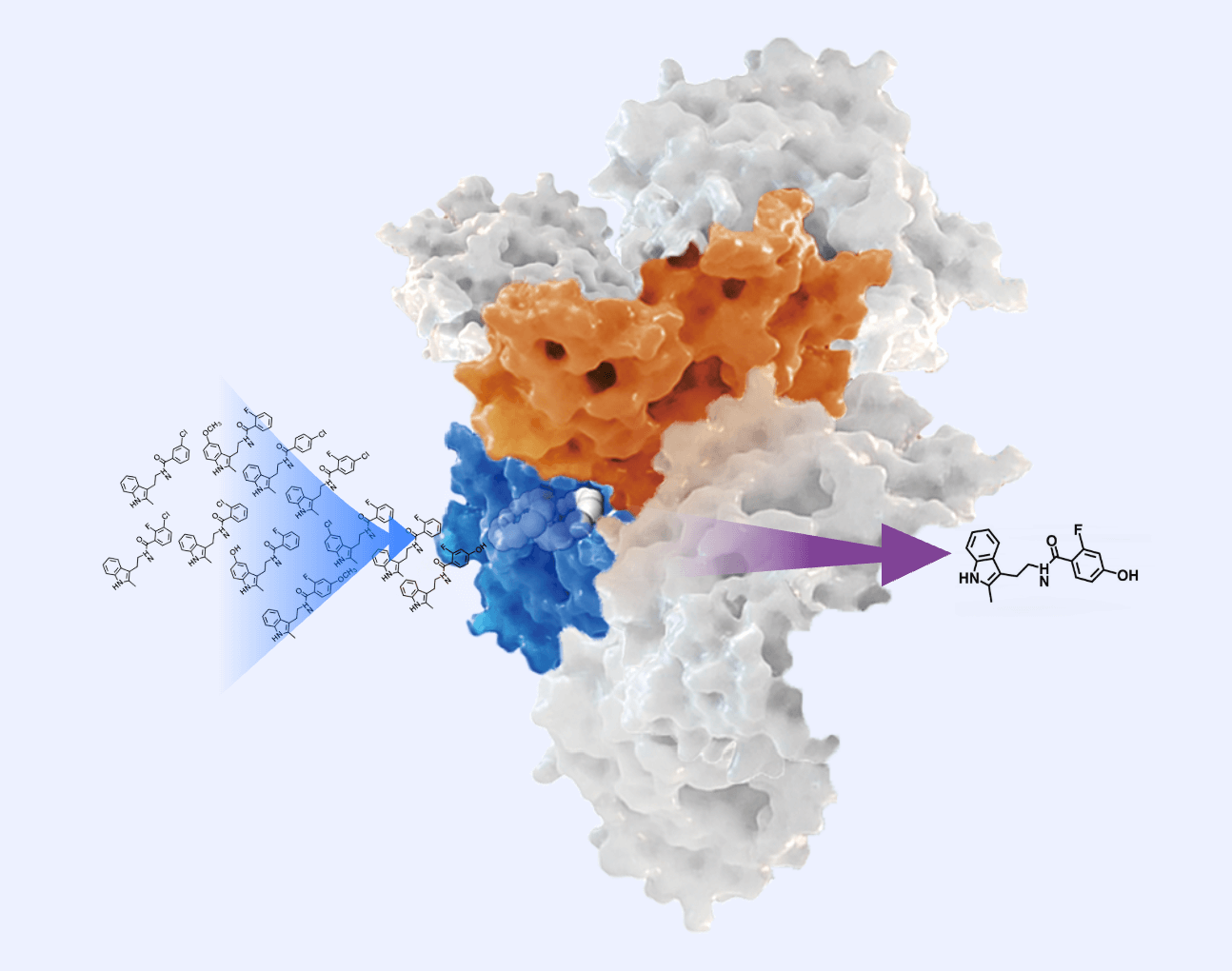
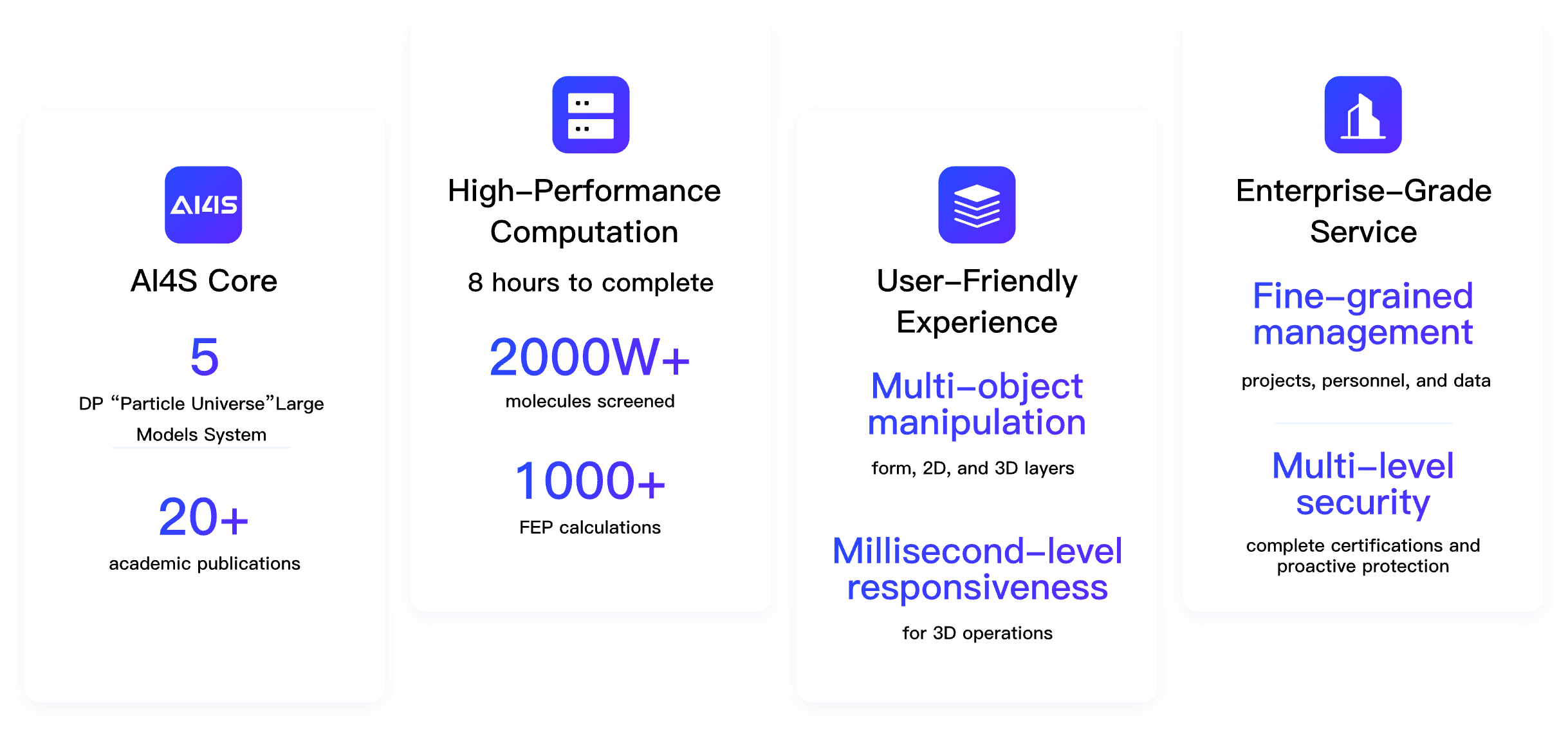
Hermite® combines artificial intelligence, physical modeling, and high-performance computing to streamline preclinical drug discovery.
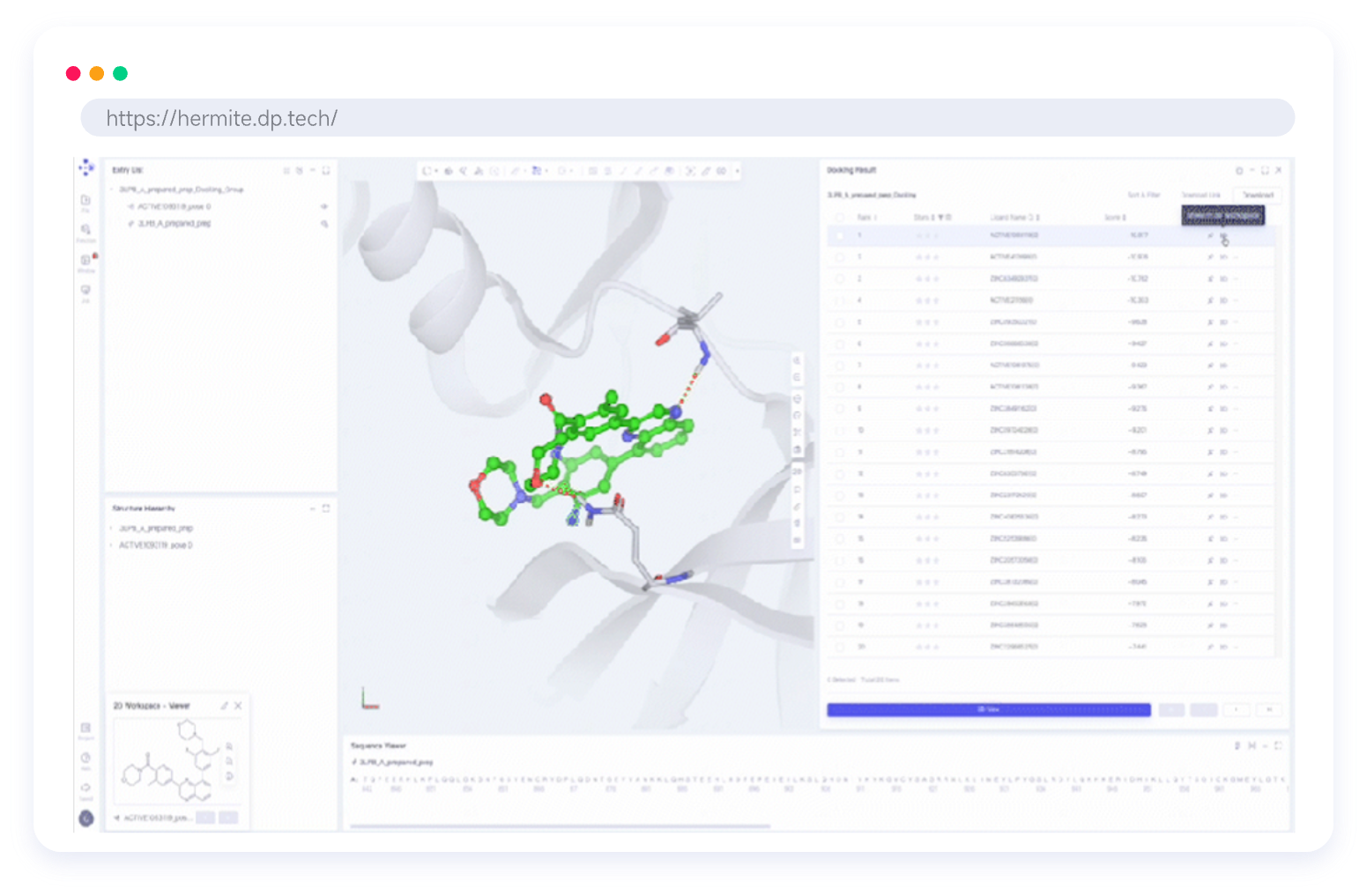
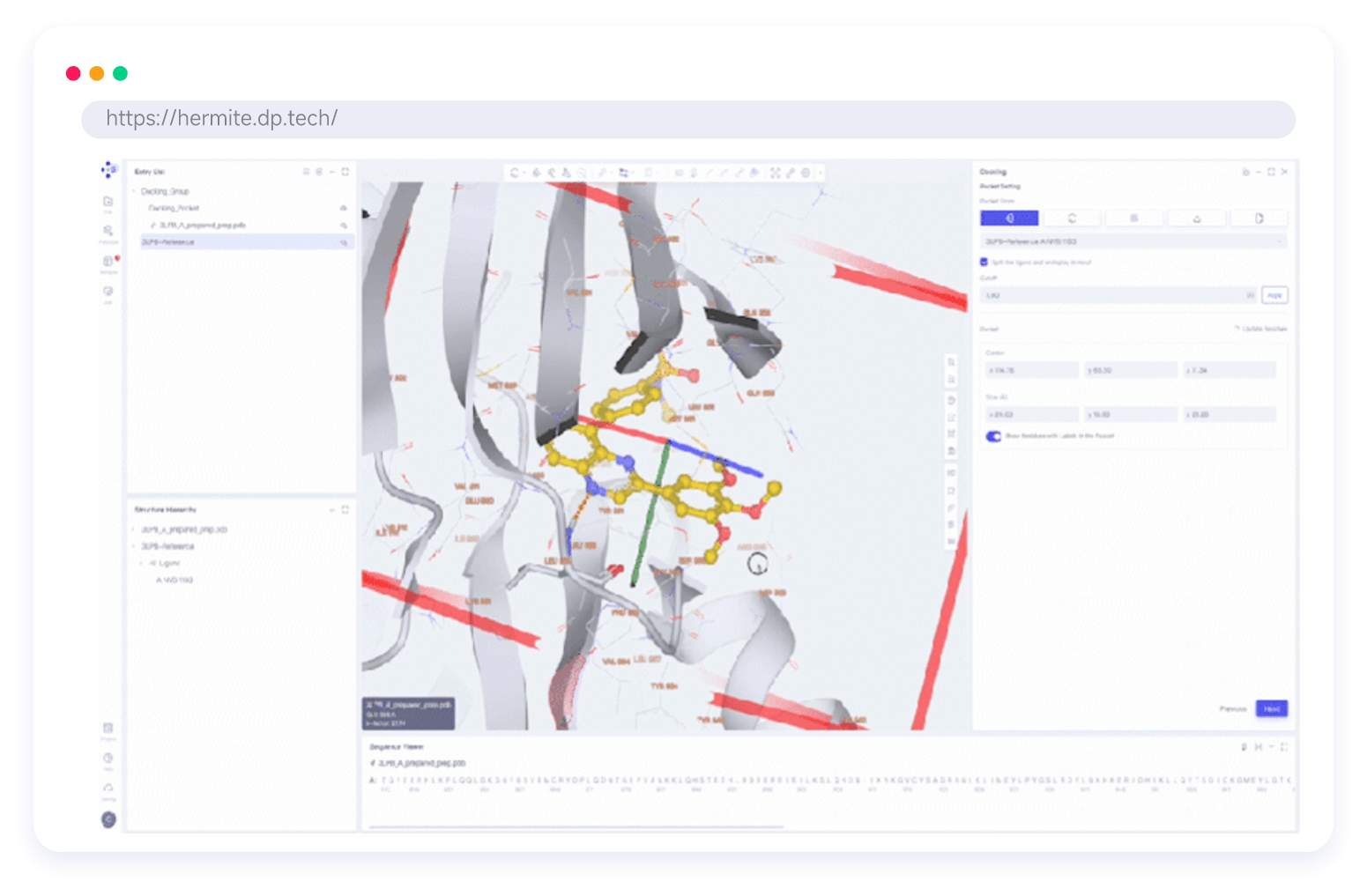
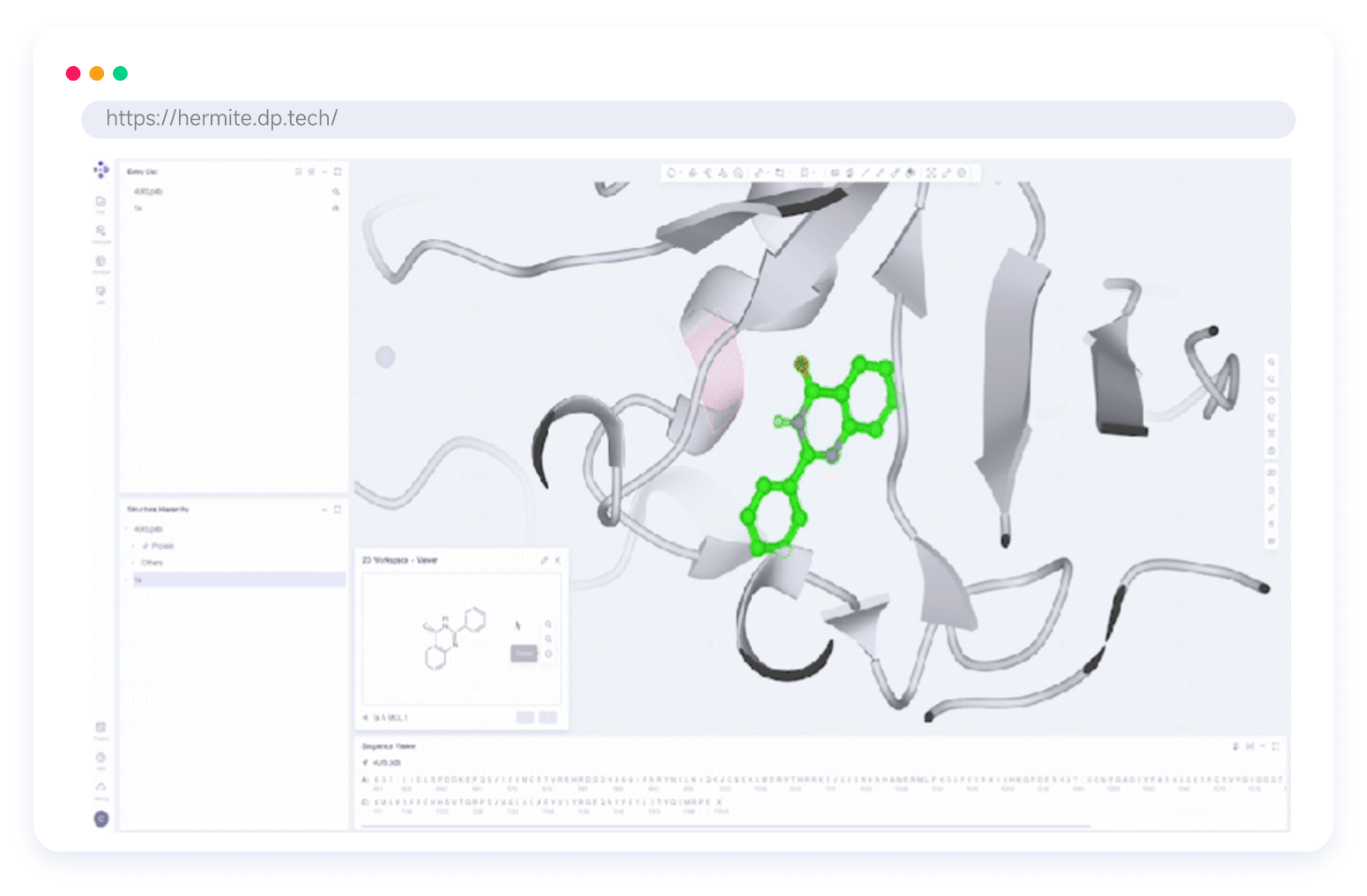
Hermite®, with massive cloud-based computational resources, supports large-scale hit compound screening and lead compound optimization, all within a single platform. Hermite® web-based molecular visualization tools enable seamless collaboration and data sharing, allowing researchers to view, design, and analyze molecular data across devices.
Custom Visualization Component Library for CADD
.png)

Security Qualifications
Certified with SOC 2 Type II, ISO 20000, ISO 27001, and ISO 9001.

System Protection
Equipped with WAF and advanced protection measures from leading cloud providers, featuring comprehensive identity verification and access control.

Dual-Insurance Mechanism
Utilizes a dual-token mechanism for data transmission and storage, ensuring robust security for user data.
Hermite® Trusted by Customers, Accelerating Pipeline Development
- Over 60% of leading pharmaceutical companies in China have chosen the Hermite® platform.
- Customers have completed more than 200,000 Hermite® Uni-FEP calculation tasks.
- The Hermite® platform has been applied in over 50 drug pipeline projects.



Partners